Next: NAMD Up: Gromacs Previous: A Box of Water
As a quick example of how to build a protein simulation system, we can consider a very recent example from the Protein Data Bank. The SARS-CoV-2 spike glycoprotein complex is an enormous protein, but a really important part of it are the domains that bind to the ACE2 receptors on the surfaces of epithelial cells. These are called Receptor Binding Domains (RBDs). A lot of recent structural biology has gone into understanding the details of the RBD-ACE2 interface. As an example, take a look at the PDB entry 7c8j, which is the X-ray crystallographic structure of a recombinant construct of the SARS-CoV-2 RBD and the ACE2 ectodomain of the bat [41]. We can use simulations to answer a lot of interesting questions about this structure, but let's just use it now as a source of coordinates to run a simulation of just the RBD alone.
The first thing to do is to download the PDB file for this entry; there are several ways to do this, but I like to use an interactive VMD session and just put 7c8j in the new molecule file browser. Once VMD has it loaded, the following TcL command in the terminal will create the stripped-down PDB file for just the RBD:
[atomselect top "chain B"] writepdb my_7c8j.pdb
Now we can pretty much follow Justin Lemkul's lysozyme tutorial here:
# generate the topology; use OPLS-AA force field (sel. 15) $ gmx pdb2gmx -f my_7c8j.pdb -o my_7c8j_processed.pdb -water spce # enlarge box $ gmx editconf -f my_7c8j_processed.pdb -o my_7c8j_newbox.gro \ -c -d 1.0 -bt cubic # solvate $ gmx solvate -cp my_7c8j_newbox.gro # copy JK's ions.mdp; add ions (replace group 13) $ gmx grompp -f ions.mdp -c my_7c8j_solv.gro -p topol.top \ -o ions.tpr $ gmx genion -s ions.tpr -o my_7c8j_solv.gro -p topol.top \ -pname NA -nname CL -neutral # minimize potential energy using JK's minim.mdp $ gmx grompp -f minim.mdp -c my_7c8j_solv.gro -p topol.top -o em.tpr $ gmx mdrun -v -deffnm em # have a look at the potential energy $ gmx energy -f em.edr -o potential.xvg # copy JK's nvt.mdp; run NVT MD for 50,000 steps -- this will take a few hours... $ gmx grompp -f nvt.mdp -c em.gro -r em.gro -p topol.top -o nvt.tpr $ gmx mdrun -v -deffnm nvt
This system has about 60,000 atoms. I show a couple of views from VMD in Fig. 36. For such a large system, we won't be able to run very long on a laptop; 100 ps takes about 3 hours on mine. Note that we'd typically want to simulate for hundreds of nanoseconds, which would be several thousand hours on my laptop, or a a day or so on a supercomputer. After about 30 ps (an hour), I went ahead and made a plot of the energies (Fig. 37).
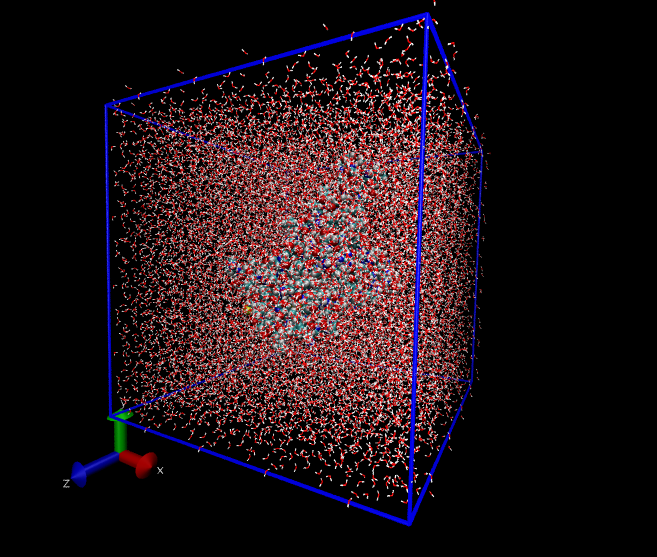
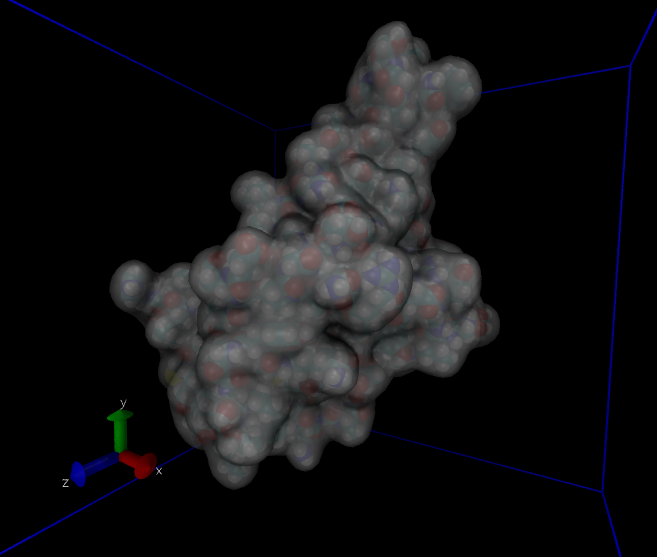 |
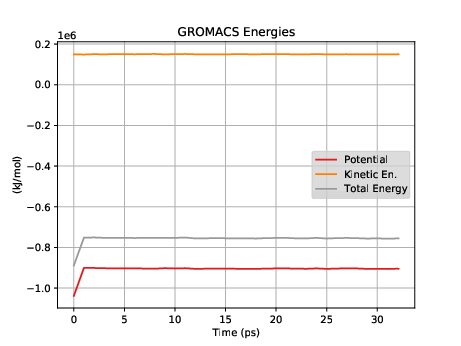 |
cfa22@drexel.edu